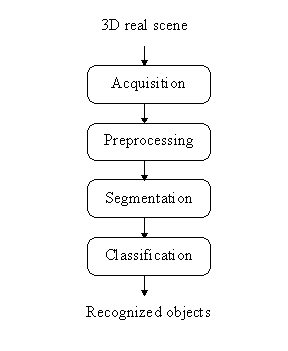
Figure 1: Typical object recognition scheme in digital image processing.
Ultrasonic imaging has become an indispensable tool in many medical examinations. A direct diagnostic value of such a method rely upon a skilled and objective interpreter of the acquired images. Therefore, assistance of a computer-based processing algorithms may play an important role on the way to a successful recognition of crucial information contained in the ultrasound recordings.
In this paper, we tackle the problem of a computer-assisted analysis of the ultrasound images of women's ovaries. As an introduction to the field, we are going, first, to talk about general principles of computerized image processing, and then, about the medical aspect of women's ovaries examination as well.
One of the frequent tasks when dealing with digital images is an object recognition or detection. Digital images are composed of big set of small rectangular elements - pixels. That is the reason why in the literature [2,3,4] the digital images are treated as grids or matrixes.
Pixels hold mostly grey level intensities (possible are also other values, e.g. textures, colours, motion gradients). The object recognition in the digital image processing sense means grouping (processing, transforming) the grey levels in such a manner that resulting groups (regions) of pixels represent objects in the image. This can be achieved on many different ways, a chart of the mostly used procedure is drawn in Figure 1.

Figure 1: Typical object recognition scheme in digital image processing.
The typical object recognition scheme has the following structure [2,3,4,7]: the first step of this scheme is an image acquisition (e.g. with the CCD camera and frame grabber). The images obtained are usually of such a poor quality (in practice the conditions of image acquisition are everything but ideal) that the extraction of the objects (using computer) only from unprocessed grey level intensity digital images is almost impossible (left image from Figure 2). Original images are because of this fact sequentially processed until object extraction is possible. Initially, there is an image preprocessing stage where the image noise found is suppressed with some filters or local operators (e.g. Gaussian, low-pass or median filter). Generally, image preprocessing enhances desirable image (object's) features. After preprocessing, the pixels which actually belong to the object have very similar values of certain features. The image segmentation follows, which can be done in many different ways, the most common methods are image segmentation based on thresholding where a few thresholds segment the entire image into regions (e.g. binary thresholding: all the pixels having a specific feature smaller than threshold form the first region, all the remaining pixels are in the other region), edge based segmentation where the object's boundary is traced, and finally, region based segmentation where the regions are searched directly (e.g. region growing or splitting). In the middle image from Figure 2 the segmentation result obtained after simple thresholding of the original image is shown.
Each obtained region is then described by some features or parameters. The last step of this scheme is classification where these regions are usually compared with praform (representative object) and after that accepted as the right object or rejected as a wrong one. The key point of a good classification is selection of appropriate features as well as a sufficiently large number of features. The right image from Figure 2 depicts recognized ellipses after classification stage.



Figure 2: Typical object recognition scheme described on an artificial image. Original noisy
image (left), two biggest regions are labeled after simple thresholding
segmentation (middle), recognized ellipses (right).
A complete understanding of follicle, i.e. a sac containing ova, dynamics inside the ovary is crucial for the field of genetic engineering. Monitoring follicles over entire cycle is especially important in human reproduction.
The outcome of a pregnancy is dependent upon the quality of the embryo. This, in turn, is dependent in part upon the quality of the female gamete oocyte contained in the dominant follicle (dominant follicles are those that grow and have potential to ovulate at the end of the follicular phase) and, therefore, the quality of the follicle itself which supports oocyte growth and maturation. Not all dominant follicles ovulate and of those that do, not all are of sufficiently high quality to result in pregnancy.
Here the main task is successfully characterize dominant follicles from the set of follicles inside the ovary. To characterize successful dominant follicles, the follicles must be compared with unsuccessful dominant and subdominant follicles and their interactions examined. For a comparison to be possible, individual large and small follicles must be identified and their development monitored over a number of days. Follicles can be monitored on many different manners, the best way of monitoring is with non-invasive methods, e.g. ultrasonography. With frames of the ovary, grabbed on either way, and with appropriate criteria (right shape, antral edge quality, size and echogenicity) the follicles (and also its type) can be identified and required analysis accomplished [1].
Today, the monitoring of follicles is done non-automatic, with human interaction. For credible results, a doctor must examine over 30 women a day during their entire cycle (ultrascan woman, freeze the ultrasound image in the best position of the ovary, measure every follicle inside ovary by hand, store the results, repeat this procedure for both ovaries - left and right). This work can be very demanding and also inaccurate.
That was the main reason that we decided to develop an application for automatic location and analysis of follicles in the ovary. In the image processing sense, we are dealing with an ultrasound image sequence of the ovary. Our work is still in the initial stage, so we are able only to present intermediate results. A core of the application is an algorithm analysing the follicle dynamics. Currently, we are analysing only one image from the sequence at a time.
The paper is organized as follows. In Section 2, our algorithm for automated detection of follicles in the ultrasound images of the ovary is described in detail, in Section 3 some results obtained are presented, followed by the section with a discussion on some drawbacks and also possible extensions of the algorithm, whereas the paper is concluded by some hints for the future work.
Before we describe the algorithm and the main ideas, let's first consider the original ultrasound image we are dealing with. There can be found the ovary with the follicles in it (the edge of the ovary is very difficult to recognize), endometrium, blood vessels and surrounding tissues. Usually, we are faced with a lot of added noise due to various reasons (e.g. produced by ultrasonic device). Our aim is at locating follicles from this original image in a step-by-step procedure that will reduce the data to be processed. The original ultrasound image of the ovary can be seen in Figure 3.
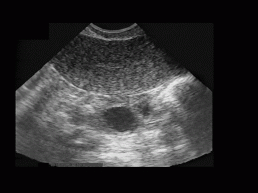
Figure 3: The
original ultrasound image of the ovary. Image dimensions are 768x576 pixels
with 256 greylevels. On the top of the image, the endometrium can be seen,
whereas the ovary with two follicles (black rounded regions) is almost
in the center of the image. The edge of the endometrium is clearly seen,
while the edge of the ovary can be located with forward and backward scanning
of the image sequence. Speckle noise and noise due to the ultrasonic device
are also noticeable (note the bright spots).
The main idea of the algorithm is to, first, coarsely estimate the ovary boundaries (resulting in a subimage containing the ovary) and, after that, to search for the follicles in this subimage. In the next subsections, the detailed description of the algorithm is given.
Our algorithm is designed according to typical object recognition scheme (Subsection 1.1), so the first step of the algorithm is image preprocessing. Because we are dealing with the ultrasound images, we must be aware of additive noise (e.g. head of the ultrascan probe is not moist enough). Especially disturbing type of noise found is speckle noise. Some authors try to reduce speckle noise with simple median filter or with combination of two median filters (with different mask sizes), but we were not satisfied with efficiency of those - so called "despeckle" filters. In paper [5] we found accurate despeckle filter called homogeneous region growing mean filter (HRGMF). This filter has one desirable feature, namely, it preserves edges. The idea of this filter is the following: for a given pixel point, a local rectangular region centered at the point is defined (initial region). If this rectangular satisfies a certain local criterion of homogeneity (this criterion can be obtained from the imaging system experimentally), it is taken as the seed region. Otherwise, the region is contracted subsequently until contracted region satisfies the homogeneity criterion. Once the seed region is determined in this way, the next step is to grow the homogeneous region by absorbing a thin, adjacent rectangular side region which has the same statistical properties as the seed region. This is repeated until there is no more homogeneous neighbour on any side of the current region. Now, the mean of the pixel values of the grown homogeneous region is mapped onto the filtering point.
The image filtered with HRGMF filter is a basis for the whole subsequent analysis. From this image we try to estimate the ovary position. This task is accomplished as follows. First, the edge detector is used. We apply Kirsh's filter for edge detection (more about the filter can be found in [2,4,7]). We experimented with a lot of other edge detecting operators (Sobel, Canny filter, LoG, etc.), but the Kirsh's one gave the most satisfying results respecting the time consumption and efficiency. Then the image is binarised (optimal threshold) and thinned. Because edges are very corrupted (ovary and follicles don't have very expressed edges), we correct them. A simple heuristic method is used for edge filling. Starting and ending point of the connected component (partial boundary) are joined (in the direction of straight line through both points) with edge pixel from the other connected component (edge pixel must be located in predefined zone near both pixels). From so processed image, the position of the ovary is estimated. The criterion used for estimation is that ovary is probably in a rectangle where the density of edges, i.e. white pixels, is the highest, since the ovary contains several regions belonging to the follicles.
The density of edges is estimated by two histograms generated along x and y direction. The maximum value in both histograms is obtained. This maximum is a starting point from which two minima, one on each side of the maximum (for each histogram), are searched for. We define minimum as the lowest value before the histogram values start to increase significantly again (this minima are not necessary the global minima on both sides). With calculation of both minima along both directions, the rectangular area probably containing the ovary is defined (subimage with the ovary).
In preprocessing phase, the rectangular area (subimage) containing the ovary was determined. With this approach (determining a subimage) the complexity of the starting problem was lowered, since not the entire image need to be searched for the follicles.
From Figure 3, we may see that follicles are of dark (almost black) circular shapes. So, our task is to find all the dark regions in the subimage and then verify if these regions could be follicles. Dark regions are obtained with thresholding the subimage. Single threshold is determined with optimal threshold selection method. Thus, the binary subimage is generated. Then, all the black regions inside the ovary are labelled (identified) and processed. Finally, each processed region is described with parameters (area, perimeter, moments, eccentricity, compactness, bounding box, etc.).
In this stage, every parametrically described region is evaluated. On the parameter basis, we decide about each region whether it is a follicle or not. At this point, additional knowledge about the problem is introduced into the algorithm (minimum and maximum size of the follicle, expected shape, etc.). Follicles are between 2 mm and 10 mm in size and they are of circular shape. This knowledge influences the predefined thresholds needed and criteria for the region classification. For the classification we used three rules: area, compactness, and eccentricity. We also experimented with some other rules, but it was evident that they were correlated.
The algorithm described in Section 2 is the core of our application called "xultra". Program "xultra" is written for an X-windows system in the programming language C using Motif1.2 libraries. It is typical menu-driven application, where also step-by-step processing can be observed (intermediate results can be displayed). In Figure 4, the main window of the application is shown. Each image is first loaded into program (submenu option "File-Open") and then sequentially processed with routines from submenu "Actions" (options "Speckle", "Kirsh", etc.) in the step-by-step processing manner or it can be processed in one single step (option "All together").
Program "xultra" (the algorithm) has been tested on the HP 715/100XC and HP 712/80 systems with 128 MB of RAM. The algorithm's computational complexity is O(n2), where n means dimensions of the image. Taking into account the images with resolution of 768x576 pixels, ovary processing and follicle detection last about 6 minutes (the majority of time is spent by despeckle filter, about two thirds).
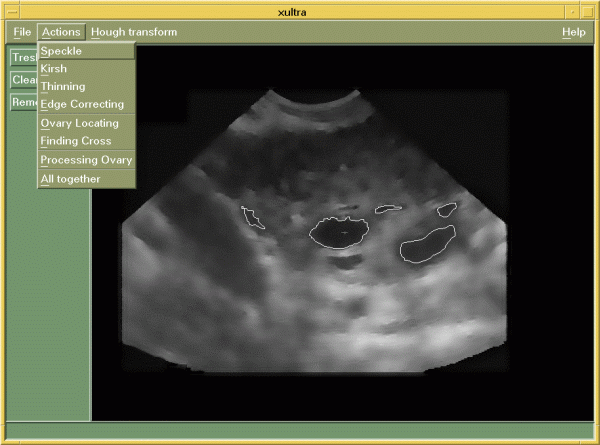
Figure 4: The main window of application "xultra".
Starting with Figure 3, the original ultrasound image of the ovary, our algorithm described in Section 2 will be demonstrated. The original images were obtained from VHS tape using simple software grabber and stored in the image format BMP with 256 greyscale palette.
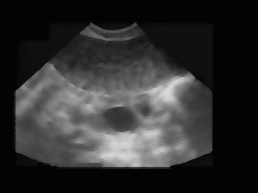
Figure 5: Figure 1 processed by HRGMF (despeckle) filter. Initial region is 7x7 pixel,
homogeneity is 20.
Foremost, this image is processed with HRGMF filter. The size of the initial region is 7x7 pixels, while homogeneity threshold is fixed to 20. This fixed values were proposed in [5]. The resulting image is depicted in Figure 5. Then, the edge detector (Kirsh's operator) is applied, followed by binarisation (optimal threshold 40) and thinning. Afterwards, the edges are filled. The result of this procedure is shown in Figure 6. From this image, the ovary location is estimated using histograms along x and y direction.
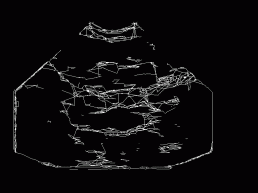
Figure 6: Kirsh's
operator applied to the despeckled image (Figure 5), followed by binarisation
(optimal threshold 40), thinning, and heuristic filling of edges.
After segmentation (Subsection 2.2), each region is classified according to the area, compactness, and eccentricity. Thresholds for classification rules are fixed and determined from the knowledge suggested by the experts. The area threshold is 220 (rule: area of follicle >220), compactness threshold is 40 (compactness > 40), and eccentricity threshold is 0.5 (eccentricity > 0.5). If a region satisfies all three criteria, then we say that this region is probably a follicle. Figure 7 depicts the recognized follicles superimposed (outlined in white) on the despeckled image. Obtained results could be compared to Figure 8, where the ovary and follicles are determined manually by a doctor.
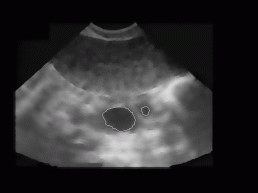
Figure 7: The
result of our current algorithm for the image from Figure 3 - recognized
follicles are rounded in white and are superimposed to the original despeckled
image. The algorithm calculates the boundary, area, perimeter, and dimensions
of the follicle along the primal axes. For the binarisation optimal threshold
(89) was used.
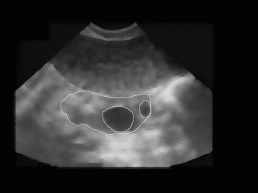
Figure 8: Ovary and two follicles as manually decided for by an expert (doctor).
All together, we tested our algorithm on 20 different ultrasound images of women's ovary (with known position of ovary and follicles). The images were obtained from 3 different women with no known reproductive problems and grabbed on different days of their menstrual period. Images were recorded with two different ultrasonic devices (in Figure 9, two different shapes of visible part of the ultrasonic images can be seen). The left image of each image pair in Figure 9 represents follicles as recognized with our algorithm, while on the right image the exact ovary and follicle position is manually drawn by an expert.
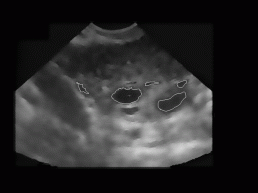
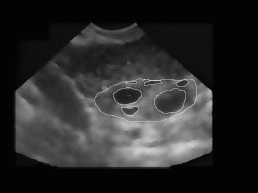
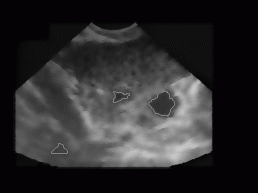
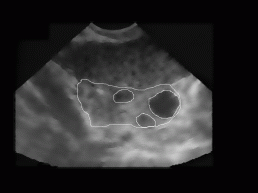
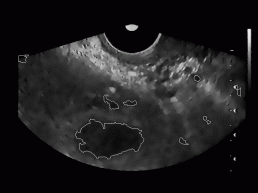
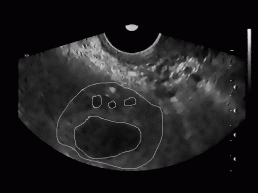
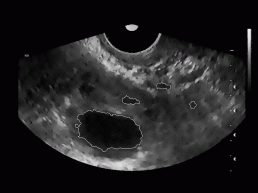
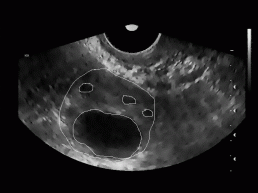
Figure 9: Some
typical results obtained with our program. On the left image of each image
pair the despeckled image with superimposed recognized follicles is depicted,
whereas on the right image the ovary and follicles as manually decided
for by an expert.
For each testing image, the ratio of correctly recognized follicles was calculated (number of follicles and their position in each image was known in advance). Besides, the average of these ratios (recognition rate of our algorithm) was obtained and was around 62%. This statistics includes recognition rate for the dominant follicles (usually are very big) as well as for the subdominant follicles (smaller than dominant follicles). Considering only the dominant follicles, the recognition rate was much higher and was around 89%.
In Figure 9, we can also notice some misidentified regions. For each testing image the misidentification rate defined as a ratio between all misidentified regions and all recognized regions in the image was determined. The average misidentification rate is around 47%.
In Section 3, we presented results obtained by our algorithm. In the next paragraphs, the algorithm will be evaluated and some improvements proposed.
First, a big drawback of the algorithm is that it requires too much processing time (about 6 minutes per image). We isolated the most time consuming part of it - HRGMF filter (which lasts about 4 minutes). This filter gives excellent results indeed, but we could also apply less efficient (with lower computational complexity) despeckle filters (e.g. geometric filter or wavelet transform), corrupted results can be, however, partly recovered in later phases (e.g. with morphological operators). Another solution which could decrease processing time is parallel image processing. Especially preprocessing stage is suitable for parallelism, where each pixel is processed in the same manner. The speedup factor can be up to number of processing units high, that means that if we had 10 processing units, the processing time would be around 10 times shorter (in our case not 4 minutes, but about 30 seconds) [6].
Our algorithm works with few predefined thresholds (e.g. at ovary estimation) which depend on type of ultrasonic device. If images obtained from another type of ultrasonic device are going to be tested, then a calibration of the required thresholds must be done.
The estimated ovary is sometimes too big, which can cause misidentification of many dark regions, very similar to follicles (by their shape, size etc.), but actually laying outside the boundary of the real ovary. We recommend inclusion of statistical analysis at this point (e.g. calculating greylevel distribution for the ovaries and follicles with known position, then applying this knowledge to the original images), by which the estimated ovary position could be verified (and corrected).
Optimal threshold method used for the subimage segmentation gives normally good results, but in some ultrasound images of the ovary it causes merging of regions (threshold too low). That means that one bigger region is generated (containing two or more neighbouring follicles) which is misidentified. Similarly, a few very small regions could be generated (threshold too high) which don't belong to the actual follicles (because of the required parameters, it is important to recognize follicles in its original size). All this indicate that more powerful segmentation method should be incorporated into the algorithm. Currently, we are experimenting with the multithresholding and region-growing segmentation methods.
All the thresholds used for classification rules were obtained experimentally. Nevertheless, not all classified regions are actually follicles (in Figure 9 some identified regions aren't follicles). This problem could be solved partially. First, a more precise ovary estimation could be required that will reduce misidentified regions outside (near) the ovary. Second, all the misidentified regions inside the ovary could be removed by additional statistical analysis of greylevels inside the regions and with a few additional rules (e.g. observing the jagged edge).
In the classification procedures also one another problem arises, namely, how to treat the regions which have their features very close to the fixed thresholds. Such cases require special attention. All the available details about regions should be carefully examined and only then the decision (whether region is a follicle or not) should be made.
Because the obtained regions don't completely fit to follicles (e.g. on one side the region is smaller than the follicle, see Figure 9), however, the data about follicles should be very precise for correct medical diagnosis, we consider to apply one of the active contour methods [2,4] in order to converge coarsely detected region boundary to the actual follicle boundary.
In this paper a new algorithm for follicle detection inside the ultrasound images of the ovary was presented. The algorithm adopts a classical object recognition scheme. The main idea of our approach is, first, to locate the ovary (coarse estimation), and then, inside the ovary to search for the follicles. Detailed algorithm description is in Section 2. The algorithm was implemented and tested on original ultrasound images. Section 3 brings up some results obtained, while in Section 4 we saw that this procedure has also some shortcomings, e.g. computational complexity. Also some new ideas for the algorithm improvement were proposed.
Our attempt to detect follicles using computer is completely new in the field of genetic engineering. This algorithm is a basis for the program which will automatically trace and analyse the follicles inside the ovary during entire female cycle. It will significantly disburden experts of their everyday routines.
Despite the proposed algorithm's improvements, which will certainly rise the recognition rate of the follicles, we also study methods for direct follicle searching in ultrasound images of the ovary. The main guidance is to find less time consuming methods.
Misidentification rate of our algorithm (non-follicle regions which are classified as the follicles) is still very high, but in Section 4 we proposed some possible algorithm extensions, which could significantly reduce this rate.
The authors would like to acknowledge a valuable contribution of Dr. M.A. Gore, Department for Reproductive Biology from German Primate Centre Göttingen, Germany, and Prof. Dr. V. Vlaisavljević, Unit for Human Reproduction and Endocrinology, Teaching Hospital Maribor, Slovenia, who kindly provided the ultrasound images of the ovaries, annotated manually the correct ovary and follicle positions in the tested set of images, and critically evaluated the results obtained by our computer-assisted recognition algorithm.
Xultra with some sample ultrasound images